On 2 October 2025, following their publication in Cell Stem Cell revealing that cells can perceive rapid stiffness changes through the accumulation of force-signalling molecules via light-controlled hydrogels, Academician Gu Xiaosong, Professor Jiang Chunping, and Assistant Researcher Yang Jiapeng, in collaboration with Professor Wei Qiang's research team from Sichuan University, have further elucidated the mechanism of cell migration in soft matrix environments. They have for the first time demonstrated that dynamic changes in soft matrix stiffness trigger a novel high-speed migration mode by inducing mechanical imbalance between intracellular and extracellular forces. This research, titled ‘Dynamic Rigidity Changes Enable Rapid Cell Migration on Soft Substrates,’ has been published in Nature Communications (Impact Factor: 15.7).

Within living organisms, cell migration is central to vital processes including tissue development, wound healing, and even cancer metastasis. Conventional wisdom holds that cells prefer migration on stiffer substrates, as soft surfaces (below 4 kPa) fail to provide sufficient traction. Yet numerous human organ tissues—such as the lungs and liver—are inherently soft and dynamically fluctuating, leaving the efficient migration of cells within such environments an enduring enigma.
Dynamic Stiffness Modulation Unlocks Cellular Migration Code on Soft Substrates
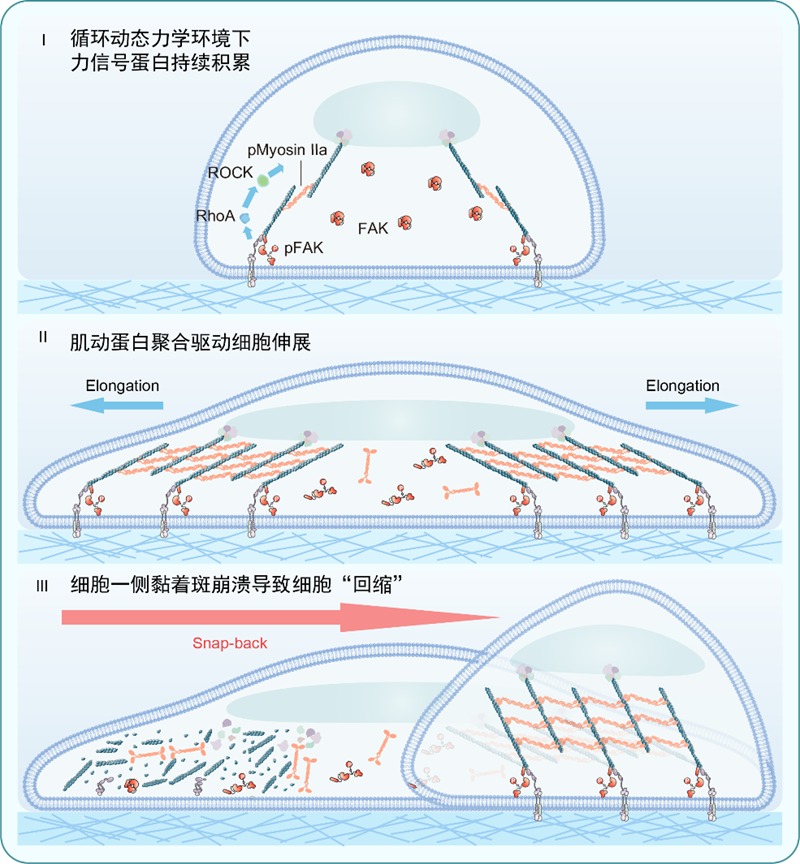
The research team employed their developed light-responsive hydrogels to construct precisely controllable dynamic mechanical environments. The study revealed that when substrate stiffness rapidly cycled between 1.6 kPa and 2.2 kPa at one-minute intervals, cell migration speed increased by over 36-fold. This surpassed traditional migration rates observed on a static 13.0 kPa rigid substrate, overturning the conventional understanding that ‘cells struggle to migrate on soft substrates’. Unlike conventional, directionally defined mesenchymal migration, cells on dynamic soft substrates exhibited a unique ‘elongation-and-snapback’ behaviour: after extending in random directions at both ends, one end rapidly snapped back, repeating this cycle continuously. This migration pattern, requiring no cell polarity establishment, demonstrated highly efficient yet non-directional ‘exploratory’ movement.
Force Imbalance Mechanism Drives Cell Migration
Previous studies revealed that dynamic stiffness cycles lead to the accumulation of force-signalling proteins, thereby enhancing internal contractile forces and driving cell extension. This research further demonstrates that the rapid increase in these internal contractile forces disrupts the equilibrium between internal and external forces. When internal contractile forces persistently exceed external adhesive forces, all focal adhesions on one side of the cell disintegrate, causing the cell body to rapidly ‘snap back’ and complete its displacement. This dynamic stiffness-induced ‘force imbalance’ constitutes the core mechanism driving migration. Experimental validation demonstrated that when force imbalance was ‘corrected’ by enhancing adhesion or inhibiting contraction, the rapid migration phenomenon ceased, robustly confirming this mechanism.
Scientific Implications:
This research not only uncovers a novel cell migration paradigm but also deepens our understanding of cellular force perception mechanisms, demonstrating that cells ingeniously exploit dynamic environmental changes to overcome static constraints. At the physiological level, this ‘search-and-settle’ migration pattern facilitates rapid exploration of microenvironments within the body—such as swiftly departing injury sites during tissue repair and subsequently ‘settling’ in stable environments. However, in pathological states, this mechanism may also be ‘exploited’ by entities such as cancer cells. Within tissues where mechanical homeostasis is disrupted (e.g., in cancer or fibrosis), this enables cells to acquire enhanced migratory capabilities, thereby increasing the risk of invasion and metastasis.
This research received funding from the Shandong Provincial Key R&D Programme (SYS202202), the Jinan Microecological Biomedicine Shandong Laboratory (JNL-2023004Q, JNL-2025008B, JNL-2025009B, JNL-2025010B), and the Shandong Provincial Natural Science Foundation (ZR2024QC217). The Jinan Provincial Laboratory served as the primary contributing institution.